About
About PICKLE
PICKLE (Protein InteraCtion KnowLedgebasE) is a meta-database for the direct protein interaction network in of human and mouse developed and maintained by:
The need for source PPI database integration and related issues
Presently, a vast insight into the known human and mouse interactome can only be achieved by means of integration of source PPI databases collecting experimental PPI data from the literature. This is due to the fact that
source PPI databases display limited overlap in their datasets due to different objectives, curation rules and subsets of the literature that they process.
PPI annotation discrepancies between the source databases:
- distinct curation rules
- different primary protein identifier types (i.e. gene, nucleotide sequence (mRNA), or protein (UniProt) IDs)
Typically, these heterogeneous primary PPI datasets are integrated via normalization: interactions are first converted to a certain target level of genetic reference and then merged.
Drawbacks:
- the node set of the integrated network is not standardized, but depends each time on the number and type of the combined sources
- different meta-databases are not directly comparable
- difficult to evaluate the expansion of the reconstructed human protein interactome over time
- possible artifacts are introduced in the integrated PPI network originating from the currently unresolved part of the human proteome
- this integration process is irreversible
- the a priori normalization approach suspends the connection between the primary and the integrated PPI networks
- it hinders the identification of normalization artifacts in the integrated network due to the inherent nonlinearity of the genetic information flow
The PICKLE approach to source PPI database integration
PICKLE introduces the concept of ontological integration as an alternative to the traditional integration via normalization.
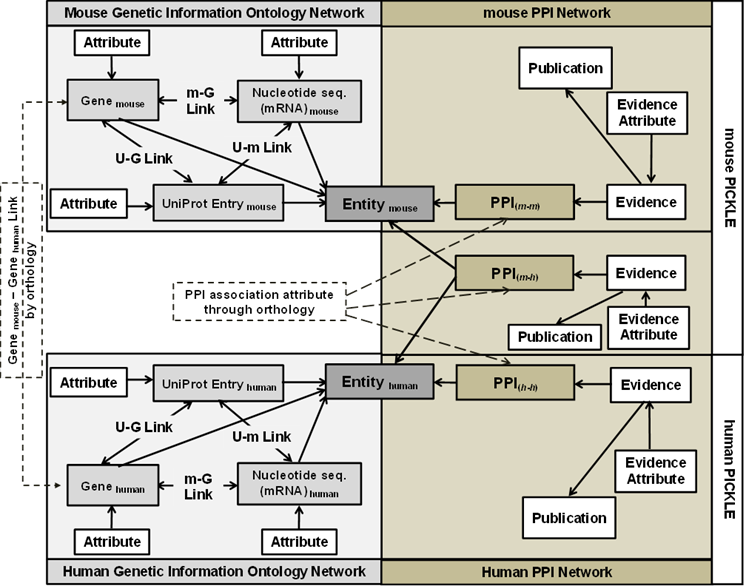
The genetic information ontology network:
- links gene, transcript and protein genetics levels
- is based on the reviewed human/mouse complete proteome (RHCP/RMCP) of UniProtKB/Swiss-Prot as a standardized reference node set
- integrates primary PPI datasets into an heterogeneous network without the need of any a priori transformations.
- allows the integrated network to be reversibly normalized to any genetic level without loss of source information
- enables primary PPI dataset cross-checking.
Focusing primarily on direct physical PPIs, PICKLE introduced a PPI filtering protocol, according to which, only PPIs with at least one supporting experiment capable of suggesting direct interactions are selected from the source PPI databases.
Moreover, PICKLE establishes an enhanced confidence scoring protocol for the PPIs being direct based on the "cross-checked" quality of the supporting experimental evidence and provides equivalent integrated human PPI networks at both the protein (UniProt) or the gene level, at three PPI filtering modes:.
- cross-checked (default)
- standard
- unfiltered
PICKLE 3.0: Upgrading PICKLE with the mouse protein interactome
PICKLE is presently upgraded with the laboratory mouse protein interactome.
Experimental PPI data between mouse genetic entities are substantially complemented by PPIs between mouse and human genetic entities.
The relational scheme of PICKLE 3.0 has been amended to exploit the Mouse Genome Informatics (MGI) mouse-human ortholog gene pair collection, enabling (i) the extension through orthology of the mouse interactome with potentially valid PPIs between mouse entities based on the experimental PPIs between mouse and human entities, and (ii) the comparison between mouse and human PPI networks.
PICKLE 3.0 provides a unique comprehensive representation of the mouse protein interactome.
The advantages of the PICKLE structural scheme
The use of the UniProtKB/Swiss-Prot defined reviewed human complete proteome (RHCP) and the reviewed mouse complete proteome (RMCP) as the basis of the instantiation of the respective PICKLE ontological networks offers a standardized point of reference between the PPI networks of successive versions and any future releases of PICKLE.
In this way, by comparing the default network of human or mouse PICKLE at the UniProt protein level between two PICKLE versions and/or subsequent releases, we can directly and reliably evaluate the manner of expansion of the RHCP or RMCP PPI network.
Moreover, the reversible normalization process achieved by PICKLE guarantees a direct correspondence between the various normalized instances of the human or mouse integrated PPI network.
Hence, one can compare a PPI in the integrated network with all the different ways in which it was originally represented in the primary PPI datasets.
This feature enables the identification of any potential normalization artifacts through the comparison of the human or the mouse protein interactome at the protein (UniProt) and gene levels and a PPI reliability assessment through cross-checking of the available experimental evidence sets contributed by the various primary datasets.
In addition, PICKLE 3.1 enables the storage of interactions between protein and gene or nucleotide sequence (mRNA) entities, e.g. those derived from protein-RNA or chromatin immunoprecipitation arrays/assays, as reported by certain sources, which can be used for purposes of cross-evaluating the supporting evidences provided in other primary datasets.
In the same context, it is possible for assorted types of data (e.g. disease-related genes, genomic, transcriptomic or proteomic data) to be consistently integrated, viewed and interpreted in the context of the protein interaction network.
Contributing Databases
PICKLE statistics - release 3.2
Statistics about the default (cross-checked) network at protein (UniProt) level regarding the primary databases:
Human PICKLE
| Database |
UniProt IDs |
PPIs |
References |
| HPRD release 9 |
9385 |
37798 |
19533 |
| IntAct release 239 |
14978 |
132539 |
9809 |
| BioGRID release 4.4.198 |
15416 |
174474 |
31429 |
| PICKLE release 3.3 |
16420 |
218025 |
45695 |
Mouse PICKLE
| Database |
UniProt IDs |
PPIs |
References |
| mouse-mouse PPI subnetwork |
|---|
| BioGRID release 4.4.198 |
4318 |
9190 |
3044 |
| IntAct release 239 |
3697 |
6037 |
2438 |
| PICKLE release 3.3 |
5666 |
13140 |
5156 |
| mouse-human PPI subnetwork |
|---|
| BioGRID release 4.4.198 |
3214 |
3488 |
1116 |
| IntAct release 239 |
3148 |
3293 |
1654 |
| PICKLE release 3.3 |
5068 |
6211 |
2626 |
The PICKLE team
|
Nicholas K. Moschonas,
Professor and Head
Department of General Biology
School of Medicine
University of Patras
Rio, Patras
&
Collaborating Faculty Member
Institute of Chemical Engineering Sciences (ICE-HT)
Foundation for Research & Technology - Hellas (FORTH)
Rio, Patras
GREECE
Phone: +30-2610-997602
e-mail: n_moschonas [a.t.] med [DOT] upatras [DOT] gr
|
Maria I. Klapa, Principal Researcher (Rank B) and Head
Metabolic Engineering and Systems Biology Laboratory (MESBL)
Institute of Chemical Engineering Sciences (ICE-HT)
Foundation for Research & Technology - Hellas (FORTH)
Rio, Patras
GREECE
Phone: +30-2610-965249
e-mail: mklapa [a.t.] iceht [DOT] forth [DOT] gr
|
|
Aris G. Gioutlakis, PhD Candidate
Department of General Biology, School of Medicine
University of Patras
&
Metabolic Engineering and Systems Biology Laboratory (MESBL)
Institute of Chemical Engineering Sciences (ICE-HT)
Foundation for Research & Technology - Hellas (FORTH)
Rio, Patras
GREECE
Phone: +30-2610-997603
e-mail: gioutlakis [a.t.] upatras [DOT] gr
|
Georgios N. Dimitrakopoulos, PhD
Department of General Biology, School of Medicine
University of Patras
&
Metabolic Engineering and Systems Biology Laboratory (MESBL)
Institute of Chemical Engineering Sciences (ICE-HT)
Foundation for Research & Technology - Hellas (FORTH)
Rio, Patras
GREECE
Phone: +30-2610-997603
e-mail: geodimitrak [a.t.] upatras [DOT] gr
|
PICKLE publications
Citing PICKLE
If you are using PICKLE, please cite:
- Dimitrakopoulos, G. N., Klapa, M. I., & Moschonas, N. K. (2022). How Far Are We from the Completion of the Human Protein Interactome Reconstruction?. Biomolecules, 12(1), 140.
- Dimitrakopoulos GN, Klapa MI, Moschonas NK. (2021). PICKLE 3.0: Enriching the human Meta-database with the mouse protein interactome extended via mouse-human orthology. Bioinformatics, 37, 1, 145-146.
- Gioutlakis A, Klapa MI, Moschonas NK. (2017). PICKLE 2.0: A human protein-protein interaction meta-database employing data integration via genetic information ontology. PLoS ONE 12(10): e0186039.
- Klapa MI, Tsafou K, Theodoridis E, Tsakalidis A, Moschonas NK. (2013). Reconstruction of the experimentally supported human protein interactome: what can we learn? BMC Syst. Biol. 7:96
PICKLE citations
Publications citing PICKLE
-
Zhou, Y., Cui, Q., & Zhou, Y. (2022). Screening and comprehensive analysis of cancer-associated tRNA-derived fragments. Frontiers in Genetics, 2893. https://doi.org/10.3389/fgene.2021.747931
-
Acharya, D., & Dutta, T. K. (2021). Elucidating the network features and evolutionary attributes of intra-and interspecific protein–protein interactions between human and pathogenic bacteria. Scientific Reports, 11(1), 1-11. https://doi.org/10.1038/s41598-020-80549-x
-
Ringwald, M., Richardson, J. E., Baldarelli, R. M., Blake, J. A., Kadin, J. A., Smith, C., & Bult, C. J. (2021). Mouse Genome Informatics (MGI): latest news from MGD and GXD. Mammalian Genome, 1-15. https://doi.org/10.1007/s00335-021-09921-0
-
Mazandu, G. K., Hooper, C., Opap, K., Makinde, F., Nembaware, V., Thomford, N. E., Chimusa E. R., Wonkam A., & Mulder, N. J. (2021). IHP-PING—generating integrated human protein–protein interaction networks on-the-fly. Briefings in bioinformatics, 22(4), bbaa277. https://doi.org/10.1093/bib/bbaa277
-
Melkonian, M., Juigné, C., Dameron, O., Rabut, G., & Becker, E. (2022). Towards a reproducible interactome: semantic-based detection of redundancies to unify protein-protein interaction databases. Bioinformatics, btac013. https://doi.org/10.1093/bioinformatics/btac013
-
Guo, X., Song, Y., Liu, S., Gao, M., Qi, Y., & Shang, X. (2021). Linking genotype to phenotype in multi-omics data of small sample. BMC genomics, 22(1), 1-11. https://doi.org/10.1186/s12864-021-07867-w
-
Panditrao, G., Ganguli, P., & Sarkar, R. R. (2021). Delineating infection strategies of Leishmania donovani secretory proteins in Human through host–pathogen protein Interactome prediction. Pathogens and disease, 79(8), ftab051. https://doi.org/10.1093/femspd/ftab051
-
Iacobucci, I., Monaco, V., Cozzolino, F., & Monti, M. (2021). From classical to new generation approaches: an excursus of-omics methods for investigation of protein-protein interaction networks. Journal of Proteomics, 230, 103990. https://doi.org/10.1016/j.jprot.2020.103990
-
Sun, F., Suttapitugsakul, S., & Wu, R. (2021). Unraveling the surface glycoprotein interaction network by integrating chemical crosslinking with MS-based proteomics. Chemical Science, 12(6), 2146-2155. https://doi.org/10.1039/D0SC06327D
-
Chen, Y., Zhou, C., Li, H., Li, H., & Li, Y. (2021). Identifying Key Genes for Nasopharyngeal Carcinoma by Prioritized Consensus Differentially Expressed Genes Caused by Aberrant Methylation. Journal of Cancer, 12(3), 874–884. https://doi.org/10.7150/jca.49392
-
Qiu, S., Munir, A., Malik, S. I., Khan, S., & Hassan, A. (2021). Identification of differentially expressed genes and pathways crosstalk analysis in Rheumatoid and Osteoarthritis using next-generation sequencing and protein-protein networks. Saudi Journal of Biological Sciences, 28(8),4656-4663. https://doi.org/10.1016/j.sjbs.2021.04.076
-
Shaukat, A. N., Kaliatsi, E. G., Skeparnias, I., & Stathopoulos, C. (2021). The Dynamic Network of RNP RNase P Subunits. International Journal of Molecular Sciences, 22(19), 10307. https://doi.org/10.3390/ijms221910307
-
Zhou, Y., Chen, H., Li, S., Chen, M. (2021). mPPI: a database extension to visualize structural interactome in a one-to-many manner, Database, Vol. 2021, baab036, https://doi.org/10.1093/database/baab036
-
Zia, J. (2021). Structural and Functional Analysis of Translocation in DISC1 gene and Impact on Schizophrenia (Doctoral dissertation, Capital University of Science & Technology , Islamabad, Pakistan).
-
Qi, Y., Guo, Y., Jiao, H., & Shang, X. (2020). A flexible network-based imputing-and-fusing approach towards the identification of cell types from single-cell RNA-seq data. BMC Bioinformatics 21, no. 1 (2020): 1-21.
https://doi.org/10.1186/s12859-020-03547-w
-
Armingol, E., Officer, A., Harismendy, O., & Lewis, E. N (2020). Deciphering cell–cell interactions and communication from gene expression. Nature Reviews Genetics.
https://doi.org/10.1038/s41576-020-00292-x
-
Singh, R. & Som, A. (2020). Role of Network Biology in Cancer Research. In: Recent Trends in ‘Computational Omics' ISBN: 978-1-53617-941-5. Editor: Pramod Katara. Nova Science Publishers, Inc.
-
Doostparast Torshizi, A., Duan, J., & Wang, K. (2020). Cell Type-Specific Proteogenomic Signal Diffusion for Integrating Multi-Omics Data Predicts Novel Schizophrenia Risk Genes. PATTERNS-D-20-00074.
http://dx.doi.org/10.2139/ssrn.3600562
-
Tao, Y. T., Ding, X. B., Jin, J., Zhang, H. B., Yang, Q. L., Chen, P. C., ... & Chen, X. (2020). HIR V2: a human interactome resource for the biological interpretation of differentially expressed genes via gene set linkage analysis. 17 June 2020, PREPRINT (Version 1) available at Research Square.
https://doi.org/10.21203/rs.3.rs-26127/v1
-
Theodosiou, T., Papanikolaou, N., Savvaki, M., Bonetto, G., Maxouri, S., Fakoureli, E., ... & Aivaliotis, M. (2020). UniProt-Related Documents (UniReD): assisting wet lab biologists in their quest on finding novel counterparts in a protein network. NAR Genomics and Bioinformatics, 2(1), lqaa005.
https://doi.org/10.1093/nargab/lqaa005
-
Koutrouli, M., Karatzas, E., Paez-Espino, D., & Pavlopoulos, G. A. (2020). A Guide to Conquer the Biological Network Era Using Graph Theory. Frontiers in Bioengineering and Biotechnology, 8, 34.
https://doi.org/10.3389/fbioe.2020.00034
-
Barreto, C. A., Baptista, S. J., Preto, A. J., Matos-Filipe, P., Mourão, J., Melo, R., & Moreira, I. (2020). Prediction and targeting of GPCR oligomer interfaces. Oligomerization in Health and Disease: From Enzymes to G Protein-Coupled Receptors, 105.
https://doi.org/10.1016/bs.pmbts.2019.11.007
-
Sadowski, M. (2020). A functional genomics approach in identifying the underlying gene for the E8 maturity locus in soybean (Glycine max) (Doctoral dissertation, Carleton University).
https://curve.carleton.ca/5c96c758-a146-4cd6-aadd-d7d83398fd3e
-
Telonis, A. G., Loher, P., Magee, R., Pliatsika, V., Londin, E., Kirino, Y., & Rigoutsos, I. (2019). tRNA Fragments Show Intertwining with mRNAs of Specific Repeat Content and Have Links to Disparities. Cancer research, 79(12), 3034-3049.
https://doi.org/10.1158/0008-5472.CAN-19-0789
-
Lin, C. Y., Lee, C. H., Chuang, Y. H., Lee, J. Y., Chiu, Y. Y., Lee, Y. H. W., ... & Wu, C. H. (2019). Membrane protein-regulated networks across human cancers. Nature Communications, 10(1), 3131.
https://doi.org/10.1038/s41467-019-10920-8
-
Wang, Y., You, Z. H., Yang, S., Li, X., Jiang, T. H., & Zhou, X. (2019). A High Efficient Biological Language Model for Predicting Protein–Protein Interactions. Cells, 8(2), 122.
https://doi.org/10.3390/cells8020122
-
Pei, J., Kinch, L., & Grishin, N. V. (2019). Mutation severity spectrum of rare alleles in the human genome is predictive of disease type. bioRxiv, 835462.
https://doi.org/10.1101/835462
-
Dimitrakopoulos, G. N., Gioutlakis, A., Klapa, M. I., & Moschonas, N. K. (2019). Evaluating the expansion of the experimentally determined human protein interactome using the PICKLE meta-database. Abstracts from the 52nd European Society of Human Genetics (ESHG) Conference: Posters. European Journal of Human Genetics, 27, 1705-1705.
https://doi.org/10.1038/s41431-019-0494-2
-
Sarkar, S., Gulati, K., Kairamkonda, M., Mishra, A., & Poluri, K. M. (2018). Elucidating Protein-protein Interactions Through Computational Approaches and Designing Small Molecule Inhibitors Against them for Various Diseases. Current topics in medicinal chemistry, 18(20), 1719-1736.
https://doi.org/10.2174/1568026618666181025114903
-
Vastrad, C., & Vastrad, B. (2018). Bioinformatics analysis of gene expression profiles to diagnose crucial and novel genes in glioblastoma multiform. Pathology-Research and Practice, 214(9), 1395-1461.
https://doi.org/10.1016/j.prp.2018.07.015
-
Wang, X., Zhai, Y., Lin, Y., & Wang, F. (2018). Mining layered technological information in scientific papers: A semi-supervised method. Journal of Information Science, 0165551518816941.
https://doi.org/10.1177%2F0165551518816941
-
Gioutlakis A., Klapa M.I., Moschonas N.K. (2018). PICKLE 2.0: A human protein-protein interaction meta-database employing data integration via genetic information ontology. ELIXIR 2018 All Hands Meeting Berlin, June 4-7 2018.
-
Steinrücken, M., Spence, J. P., Kamm, J. A., Wieczorek, E., & Song, Y. S. (2018). Model‐based detection and analysis of introgressed Neanderthal ancestry in modern humans. Molecular ecology, 27(19), 3873-3888.
https://doi.org/10.1111/mec.14565
-
Telonis, A. G., & Rigoutsos, I. (2018). Race disparities in the contribution of miRNA isoforms and tRNA-derived fragments to triple-negative breast cancer. Cancer research, 78(5), 1140-1154.
https://doi.org/10.1158/0008-5472.CAN-17-1947
-
Macalino, S., Basith, S., Clavio, N., Chang, H., Kang, S., & Choi, S. (2018). Evolution of In Silico Strategies for Protein-Protein Interaction Drug Discovery. Molecules, 23(8), 1963.
https://doi.org/10.3390/molecules23081963
-
Bojadzic, D., & Buchwald, P. (2018). Toward small-molecule inhibition of protein–protein interactions: General aspects and recent progress in targeting costimulatory and coinhibitory (immune checkpoint) interactions. Current topics in medicinal chemistry, 18(8), 674-699.
https://doi.org/10.2174/1568026618666180531092503
-
Rujirapipat, S., McGarry, K., & Nelson, D. (2017). Bioinformatic Analysis Using Complex Networks and Clustering Proteins Linked with Alzheimer’s Disease. In Advances in Computational Intelligence Systems (pp. 219-230). Springer, Cham.
https://doi.org/10.1007/978-3-319-46562-3_14
-
Papanikolaou, N., Pavlopoulos, G. A., Theodosiou, T., & Iliopoulos, I. (2015). Protein–protein interaction predictions using text mining methods. Methods, 74, 47-53.
https://doi.org/10.1016/j.ymeth.2014.10.026
-
Song, Y., & Buchwald, P. (2015). TNF superfamily protein-protein interactions: feasibility of small-molecule modulation. Current drug targets, 16(4), 393-408.
https://www.ingentaconnect.com/content/ben/cdt/2015/00000016/00000004/art00013
-
Makris, C., & Theodoridis, E. (2015). Computational Methods for Modeling Biological Interaction Networks. Pattern Recognition in Computational Molecular Biology: Techniques and Approaches, 505-524.
https://doi.org/10.1002/9781119078845.ch26
-
Song, Y. (2015). Small-Molecule Probes for the Modulation of Ligand-Receptor Interactions within the TNF Superfamily. OpenAccess Dissertations. 1398.
https://scholarlyrepository.miami.edu/oa_dissertations/1398
-
Nastou, K. C., Tsaousis, G. N., Kremizas, K. E., Litou, Z. I., & Hamodrakas, S. J. (2014). The human plasma membrane peripherome: visualization and analysis of interactions. BioMed research international, 2014.
http://dx.doi.org/10.1155/2014/397145
-
Wu, P. J., Wu, W. H., Chen, T. C., Lin, K. T., Lai, J. M., Huang, C. Y. F., & Wang, F. S. (2014). Reconstruction and analysis of a signal transduction network using HeLa cell protein–protein interaction data. Journal of the Taiwan Institute of Chemical Engineers, 45(6), 2835-2842.
https://doi.org/10.1016/j.jtice.2014.07.006
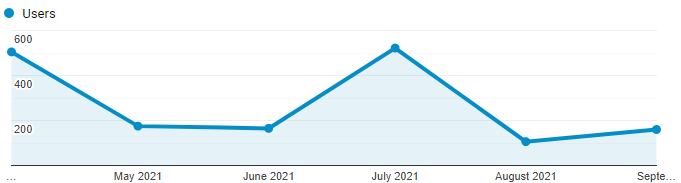
PICKLE usage
Last 6 months statistics according to Google Analytics: