Help
A. Description of the PICKLE querying web interface
The online querying interface of PICKLE comprises three parts:
i. Entity identification section
Initially, the user select the species (human or mouse) and then enters one or more protein identifiers (almost any identifier type can be used, from UniProt, nucleotide sequence (mRNA) and gene accessions or names to GO terms and chromosomal regions).
The system maps those identifiers to entities of the PICKLE RHCP/RMCP (reviewed human/mouse complete proteome) genetic information ontology network and returns the candidate list to the user for review.
ii. Interaction querying and result section
Having selected at least one entity from the list, the user can proceed and set the PPI search criteria, namely:
a) the PICKLE network normalization level (i.e. Protein (UniProt) or Gene), b) the PICKLE network filtering mode, i.e. none, standard or cross-checking (default) - see PPI filtering ruleset in next section -, and c) the PPIs which the system will retrieve from the respective PICKLE network.
Currently, the user can search for:
a) the interactions of the queried biological entities, retrieving thus their “first neighbors”, or b) the interactions of the queried entities and any interactions that may exist between those first neighbors, or c) the interactions, if any, exclusively between the queried entities.
iii. PPI network display & visualization section
The resulting interacting pairs are formatted in such a way that the first displayed interactor is always a queried entity. The interactions are sorted based on their cross-checked confidence score of a PPI, their standard confidence score (see PPI confidence scoring scheme in next section) and the total number of supporting publications, the links of which are also provided.
The user can download the resulting PPI set in a tab-delimited format and/or visualize it if its size permits (currently, less than 500 PPIs)) via the Cytoscape.js visualization plugin. The user can also retrieve the source information regarding the identified PPIs as this is recorded with the primary datasets along with the evaluation of the quality of the supporting experimental evidences ('details' link).
iv. Mouse queries and ortholog entities
In the case of mouse queries, experimental evidence might include mouse-human PPIs (PPI(m-h)). These are displayed in the results table under the mouse-mouse PPIs (PPI(m-m)).
Moreover, the orthologous pair of nodes are displayed at the end of table based on MGI homology data.
The "Linked PPI" column displays any PPI(m-h) having a corresponding PPI(m-m) based on the MGI mouse-human othology data.
The "Human PPI" column indicates any PPI(m-m) or PPI(m-h) having a corresponding PPI in the human PICKLE interactome.
The experimentally determined PPI network is shown in the visualization page. Mouse and human nodes are shown in green and orange, respectively. PPI(m-h) edges are displayed with different shades of brown, while mouse-human ortholog node pairs are grouped in a supernode.
The extended by orthology (EbO) PPI network can be displayed by pressing the corresponding button.
In the EbO PPI network, ortholog nodes are merged and PPI(m-h) are converted to PPI'(m-m) shown with a dashed line.
Thicker lines indicate duplicate PPIs, i.e. PPI(m-m) and PPI'(m-m).
B. The PICKLE experimental evidence quality evaluation and PPI confidence level scoring schemes
i. Experimental evidence quality evalution scheme
In PICKLE, what we call ‘evidence’ is an evaluated set of attributes provided by a source database regarding a specific PPI.
As of "first-class" standard quality are evaluated the sets of evidence attributes that suggest a direct protein-protein interaction.
This category includes all evidence sets, in which the interaction type (if provided by a source database) indicates a direct interaction.
In these cases, we trust the respective designation of the source database curators, regardless of the rest of the supporting experimental data, i.e. the experimental system and/or detection method and the throughput (in the case of BioGRID and DIP) or expansion method (in the case of MIntAct).
Even in the case of other interaction types, a set of evidence attributes is still evaluated as “first-class”, if the description of the supporting experimental setup accounts for methodologies capable of detecting direct interactions with a high degree of confidence (e.g. a yeast two-hybrid system).
As of "second-class" standard quality are evaluated the sets of evidence attributes that do not belong to the "first-class" category and do not describe a high-throughput experiment (in BioGRID and DIP) or do not depict that the supported PPIs have been derived from spoke expansion (in MIntAct)
The “second-class” evidence sets may suggest direct protein-protein interactions, but are subject to questioning and can be downgraded to a lower class designation given contrary information from other databases (i.e. database cross-checking).
As of "third-class" standard quality are evaluated the sets of evidence attributes that do not belong to the "first-class" category and describe a high-throughput experiment (in BioGRID and DIP) or depict that the supported PPIs have been derived from spoke expansion (in MIntAct)
In the case of the "third-class" evidence attributes, the described protein-protein interactions would be considered of low probability of being direct.
As "fifth-class" are evaluated the sets of evidence attributes that refer to interactions between proteins and genes or nucleotide sequences (mRNAs) .
Protein-gene or protein-nucleotide sequence (mRNA) interactions are stored in PICKLE for primary PPI dataset cross-checking purposes only and are not included
in the PICKLE PPI network. All genetic interference interactions (either in terms of interaction type or supporting detection method) in MIntAct, BioGRID or DIP are a priori discarded.
ii. The PPI confidence level scoring scheme
The sets of evidence attributes associated with primary PPIs assist in the definition of a standard scoring protocol for the confidence level of a PPI being direct.
Employing ontological PPI integration, PICKLE enables the cross-checking between the primary PPI datasets in the cases of curation overlaps, leading to further refinement of the standard PPI confidence scoring protocol.
In all cases, “first-class” evidences retain their classification despite any potential discord between sources. However, “second-class” evidences provided from a particular database can be downgraded in the presence of alternative, lower-class (i.e. third or fifth) evaluations of the evidences provided from other sources for the same publication. Through this process, we assign two quality evaluations to each evidence: its original ("standard") and its "cross-checked" class.
Accordingly, each interaction is given two confidence scores; the "cross-checked" confidence score of a PPI is based on the "cross-checked" quality class of its supporting evidences.
The “first” standard PPI confidence threshold assigned a standard confidence score ‘1’ includes all PPIs supported by at least one evidence set of standard “first-class” quality.
These PPIs are of high probability being direct, not subject to primary PPI dataset cross-checking; by definition their cross-checked confidence score will be '1' too.
The “second” standard PPI confidence threshold assigned a standard confidence score ‘2’ includes all PPIs supported by at least one evidence set of standard “second-class” quality and none of standard "first-class" quality.
These PPIs are of moderate probability being direct and are subject to primary PPI dataset cross-checking. Their "cross-checked" confidence score may remain
the same, if not 'challenged' during the primary PPI dataset cross-checking process or downgraded to the third or fifth "cross-checked" confidence level, if ended being supported only by third or fifth "cross-checked" quality class evidences.
The “third” standard PPI confidence threshold assigned a standard confidence score ‘3’ includes all PPIs supported by at least one evidence set of standard “third-class” quality and none of standard "first" or "second" class.
These PPIs are considered of low probability of being direct. Their cross-checked confidence score may be downgraded to the "fifth-class", if ended being supported only by
fifth "cross-checked" quality class evidences.
The “fifth” standard PPI confidence threshold assigned a standard confidence score ‘5’ includes all PPIs supported only by "fifth-class" evidences. .
These protein-gene or protein-nucleotide sequence (mRNA) interactions are stored in PICKLE for primary PPI dataset cross-checking purposes only and are not included
in the PICKLE PPI network, thus they are not displayed in the PICKLE web-based querying interface. They are included in the 'Full' interaction datasets in the Downloads section.
C. The PICKLE PPI filtering modes
Human PICKLE
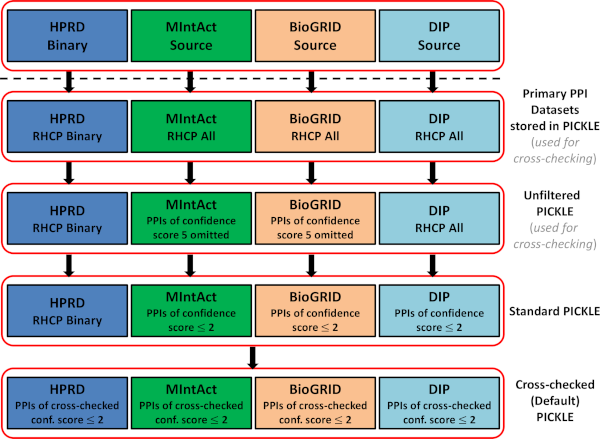
The HPRD, MintAct, BioGRID and DIP datasets included in the unfiltered, standard and cross-checked (default) PICKLE 2.0 PPI network are shown in the figure below. RHCP stands for the reviewed human complete proteome. The primary datasets stored in PICKLE involve solely associations supported by at least one experimental evidence set with linked publication(s), with both interactors designated as human and belonging to the PICKLE RHCP genetic information ontology network (available as 'Full' dataset in the Downloads section).
Mouse PICKLE
The same approach is followed in mouse PICKLE, except that only MIntAct, BioGRID and DIP datasets are used.
D. PICKLE manual and video tutorials
To acquaint yourself with PICKLE, you may download the
PICKLE manual.
Alternatively, you may watch video tutorials for the following website sections:
1. PPI Query and Results sections
2. PPI Network Visualization window
3. Downloads page
1. Video Tutorial for the PPI Query and Results sections
2. Video Tutorial for the PPI Network Visualization Window
3. Video Tutorial for the Downloads page